Evolution of metabolic pathways
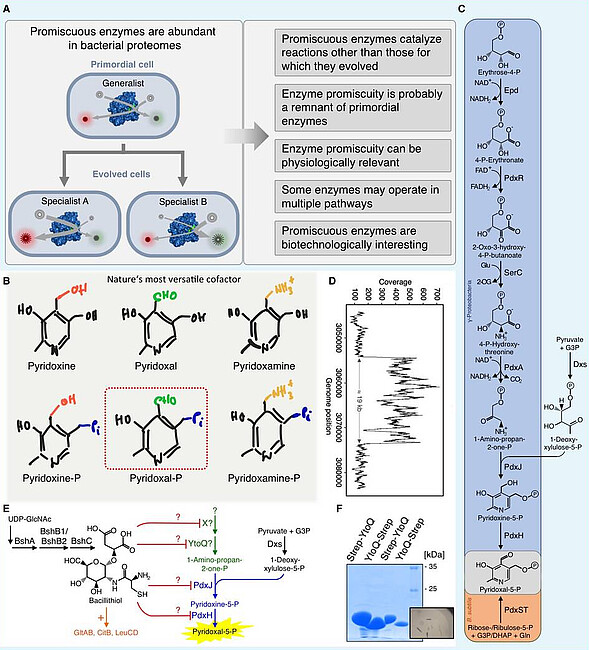
Complex bacteria like the Gram-negative and Gram-positive model organisms Escherichia coli and Bacillus subtilis, respectively, have versatile metabolic properties allowing them to thrive in a variety of niches. Due to their metabolic adaptability, these bacteria are also often used for biotechnological applications. The enzyme equipment of a cell largely determines its metabolic properties. E.g., a bacterium is only capable of growing with a certain sugar as the sole source of carbon if the cells possess a transport system and enzymes for energy conversion and for generating essential building blocks, which are required to build up a cell. Therefore, the emergence of catabolic and anabolic routes is a crucial step for the development and evolution of complex cellular systems. In the past years, several evolutionary mechanisms contributing to metabolic diversity have been proposed and experimentally validated. More than 40 years ago, it has been suggested that primordial organisms contained small genomes encoding a small number of proteins and enzymes. Furthermore, it has been proposed that the ancient enzymes of primordial cells were multifunctional with broad substrate specificities allowing the cells to carry out all metabolic reactions that are required for life (Figure A). Primordial cells may have then further evolved by gene duplication and subsequent specialization of the genes due to the accumulation of mutations facilitating metabolic diversification (Figure A). The finding that many “modern” enzymes are promiscuous supports the model that the versatile metabolic properties of extant bacteria might be due to specialization of primordial enzymes. The promiscuous enzymes that are present in extant bacteria like E. coli and B. subtilis represent a rich and almost unexplored source for the identification of biocatalysts, which might be improved and harnessed to engineer bacteria for biotechnological applications such as the degradation of anthropogenic substances (Figure A). It has been estimated that about 37% of the E. coli enzymes act on several substrates and catalyse 65% of the known metabolic reactions. Thus, the interconnection of metabolic pathways via promiscuous enzymes is probably much more common than previously anticipated. Recently, we reported on the existence of promiscuous enzymes in B. subtilis that convert two non-natural substrates into vitamin B6, which is of commercial interest. Moreover, B. subtilis possesses at least two enzymes that can functionally replace each other in the threonine biosynthetic pathway. It has also been shown for E. coli that a generalist enzyme contributes with physiologically relevant levels to two distinct pathways. However, even though E. coli and B. subtilis have been intensively studied during the last decades, the degree of enzyme promiscuity in these bacteria remains to be experimentally validated. We are interested in vitamin B6 metabolism in bacteria. Vitamin B6 is a designation for the six vitamers pyridoxal, pyridoxine, pyridoxamine, pyridoxal 5′-phosphate (PLP), pyridoxine 5′-phosphate, and pyridoxamine 5′-phosphate (Figure B). PLP, being the most important B6 vitamer, serves as a cofactor for many proteins and enzymes (B6 Database). In contrast to other organisms like bacteria, animals and humans have to ingest vitamin B6 with their food. Therefore, vitamin B6 is a valuable compound for the pharmaceutical and the food industry. Two metabolic pathways for de novo synthesis of PLP have been described (Figure C). The long pathway is present in proteobacteria like the Gram-negative model bacterium Escherichia coli and depends on the phosophosugar deoxyxylulose 5-phosphate (DXP). In the DXP-dependent vitamin B6 pathway, seven enzymes are required for PLP synthesis. The short and DXP-independent vitamin B6 biosynthetic pathway is present in plants, fungi and in the Gram-positive bacteria like Bacillus subtilis. The short pathway involves the PdxST PLP synthase enzyme complex. Recently, it has been shown that a block in the DXP-dependent vitamin B6 pathway in E. coli can be bypassed by the overproduction of promiscuous enzymes. Thus, a functional pathway can be patched together from native promiscuous enzymes without mutations in the coding genes. We have observed that the introduction of the last part of the DXP-dependent vitamin B6 pathway into a B. subtilis pdxST mutant strain lacking the PdxST PLP synthase enabled the bacteria to evolve and grow in the absence of exogenously provided PL. The evolved strain overproduce the YtoQ gene of unknown function by selective gene amplification (Figure D). Moreover, all evolved strains have inactivated gene, which are involved in the low-molecular weight thiol bacillithiol (Figure E). The underlying molecular mechanism allowing the bacteria to produce vitamin B6 at physiologically relevant levels is currently under investigation. We are also interested in identifying the function of YtoQ (Figure F). However, the so-called underground metabolism or side activities of prevalent metabolic enzymes enable the bacteria to make use of native and non-native enzymes for metabolic rewiring.
Selected publications
Mardoukhi MSY, Rapp J, Irisarri I, Gunka K, Link H, Marienhagen J, de Vries J, Stülke J, Commichau FM (2024) Metabolic rewiring enables ammonium assimilation via a non-canonical fumarate-based pathway. Microb Biotechnol. 17: e14429.
Stecker D, Hoffmann T, Link H, Commichau FM, Bremer E (2022) L-Proline synthesis mutants of Bacillus subtilis overcome osmotic sensitivity by genetically adapting L-arginine metabolism. Front Microbiol. 13: 908304.
Richts B Lentes S Poehlein A Daniel R Commichau FM (2021) A Bacillus subtilis pdxT mutant suppresses vitamin B6 limitation by acquiring mutations enhancing pdxS gene dosage and ammonium assimilation. Environ Microbiol Rep. 13: 218-233.
Richts B, Commichau FM (2021) Underground metabolism facilitates the evolution of novel pathways for vitamin B6 biosynthesis. Appl Microbiol Biotechnol. 105: 2297-2305.
Acevedo-Rocha CG Gronenberg LS Mack M Commichau FM Genee HJ (2019) Microbial cell factories for the sustainable manufacturing of B vitamins. Curr Opin Biotechnol 56: 18-29.
Richts B Rosenberg J Commichau FM (2019) A survey of pyridoxal 5′-phosphate-dependent proteins in the Gram-positive model bacterium Bacillus subtilis. Front Mol Biosci 6: 32.
Rosenberg J Commichau FM (2019) Harnessing underground metabolism for pathway development. Trends Biotechnol 37: 29-37.
Rosenberg J Yeak KC Commichau FM (2018) A two-step evolutionary process establishes a non-native vitamin B6 pathway in Bacillus subtilis. Environ Microbiol 20: 156-168.
Rosenberg J Ischebeck T Commichau FM (2017) Vitamin B6 metabolism in microbes and approaches for fermentative production. Biotechnol Adv 35: 31-40.
Rosenberg J Müller P Lentes S Thiele MJ Zeigler DR Tödter D Paulus H Brantl S Stülke J Commichau FM (2016) ThrR, a DNA-binding transcription factor involved in controlling threonine biosynthesis in Bacillus subtilis. Mol Microbiol 101: 879-893.
Commichau FM Alzinger A Sande R Bretzel W Reuß DR Dormeyer M Chevreux B Schuldes J Daniel R Akeroyd M Wyss M Hohmann H Prágai Z (2015) Engineering Bacillus subtilis for the conversion of the antimetabolite 4-hydroxy-L-threonine to pyridoxine. Metab Eng 29: 196-207.
Commichau FM Alzinger A Sande R Bretzel W Meyer FM Chevreux B Wyss M Hohmann H Prágai Z (2014) Overexpression of a non-native deoxyxylulose-dependent vitamin B6 pathway in Bacillus subtilis for the production of pyridoxine. Metab Eng 25: 38-49.
