Phage biology
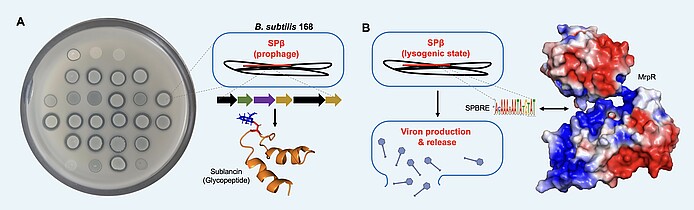
Phages are bacterial viruses whose replication depends on the host cell. Most host cells are killed after phage replication. Some phages may enter the lysogenic state or temperate life cycle. When a phage enters the lysogenic life cycle, the phage genome is integrated into the chromosome of the host cell and the bacteria carrying the prophage become lysogen. Prophages that multiply together with the host chromosome may enter the lytic cycle through the action of DNA-damaging agents. We are interested in the biology of the temperate phage SPβ that infects the Gram-positive model bacterium Bacillus subtilis. SPβ was discovered about 50 years ago and resembles the Siphoviridae morphotype [Kohm and Hertel, 2021; Kohm et al., 2022, 2023a]. SPβ is a benefitting trait after the transfer to another B. subtilis strain because cultures of the bacterium carrying the prophage release the S-linked glycopeptide sublancin, which belongs to the class of antimicrobial natural products named glycocins. Sublancin inhibits the growth of B. subtilis strains that do not carry SPβ (Figure A) [Garcia de Gonzalo et al., 2015]. In this project, we study the lysis/lysogeny management system of SPβ that is required for maintaining the lysogenic state of the prophage and also allows the entry into the lytic cycle. Recently, we discovered the repressor MrpR that prevents SPβ from entering the lytic cycle by binding to the SPbeta repeated (SPBR) element (Figure B) [Kohm et al., 2023b]. The SPBR element was described 25 years ago but the protein recognizing the DNA sequences remained so far elusive [Lazarevic et al., 1999]. We employ genetic as well as biochemical methods to elucidate the functions of key components of the lysis/lysogeny management system.
Selected publications
Kohm K, v. Clanner A, Hertel R, Commichau FM (2025) Closely related and yet special - how SPβ phages control lysis-lysogeny decisions. Trends Microbiol. 33: 387-396.
Kohm K, Jalomo-Kharyova E, Krüger A, Basu S, Steinchen W, Bange G, Frunzke J, Hertel R, Commichau FM, Czech L (2023b) Structural and functional characterization of MrpR, the master repressor of the Bacillus subtilis prophage SPβ. Nucleic Acids Res. 51: 9452-9474.
Kohm K, Lutz VT, Friedrich I, Hertel R (2023a) CRISPR-Cas9 shaped viral metagenomes associated with Bacillus subtilis. Methods Mol Biol. 2555: 205-212.
Kohm K, Floccari VA, Lutz VT, Nordmann B, Mittelstädt C, Poehlein A, Dragos A, Commichau FM, Hertel R (2022) The Bacillus phage SPβ and its relatives: a temperate phage model system reveals new strains, species, prophage integration loci, conserved proteins and lysogeny management components. Environ Microbiol. 24: 2098-2118.
Kohm K, Hertel R (2021) The life cycle of SPβ and related phages. Arch Virol 166: 2119-2130.
Garcia de Gonzalo CV, Denman EL, Mars RA, Stülke J, van der Donk WA, van Dijl JM (2015) The phosphoenolpyruvate:sugar phosphotransferase system is involved in sensitivity to the glucosylated bacteriocin sublancin. Antimicrob Agents Chemother. 59: 6844-6854.
Commichau FM, Stülke J (2012) A mystery unraveled: essentiality of RNase III in Bacillus subtilis is caused by resident prophages. PLoS Genet 8: e1003199.
Lazarevic V, Düsterhöft A, Soldo B, Hilbert H, Mauel C, Karamata D (1999) Nucleotide sequence of the Bacillus subtilis temperate bacteriophage SPbetac2. Microbiology 145: 1055-1067.