Evolution of herbicide resistance
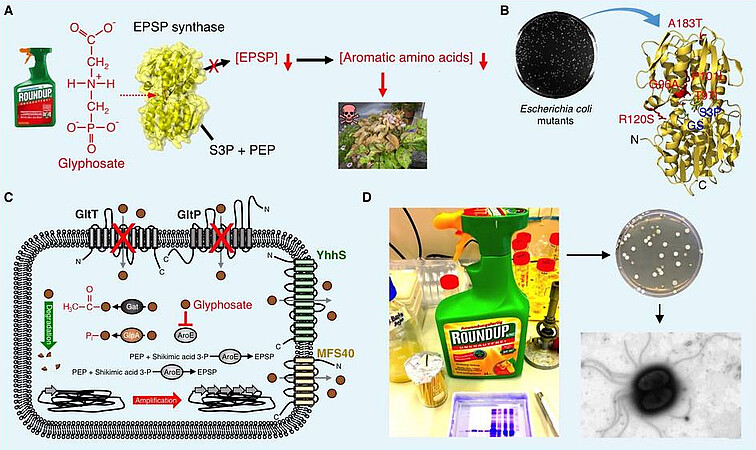
Bacillus subtilis is a soil bacterium that can get into contact with herbicides. Glyphosate (GS) is a non-selective herbicide that kills weeds and other plants that compete with crops. GS inhibits the 5-enolpyruvyl-shikimate-3-phosphate (EPSP) synthase in plants and other organisms including bacteria [Hertel et al., 2021]. The EPSP synthase converts phospoenolpyruvate (PEP) and shikimic acid-3-phosphate (S3P) into EPSP serving as an essential precursor for de novo synthesis of the aromatic amino acids, folates and quinones (Figure A). Therefore, the inhibition of the EPSP synthase by GS results in cell death (Figure A). Few years after the discovery of the mode of action of GS, several GS-insensitive EPSP synthase variants have been generated by either random or directed mutagenesis or they have been isolated from GS-resistant organisms (Figure B). The structural characterization of EPSP synthases revealed that GS targets the PEP binding site and acts as a competitive inhibitor of the enzyme. EPSP synthases that are insensitive to GS have been used to genetically engineer GS-tolerant crops. Even though B. subtilis is a well-studied organism, the information about the interaction between this bacterium and GS as well as its potential to adapt to the herbicide was limiting. Using B. subtilis as a model organism, we identified the first GS transporters (Figure C) [Wicke et al., 2019]. Serendipitously, we isolated bacteria from a commercially available Roundup® solution (Figure D). The bacteria belong to the genus Burkholderia and tolerate high amounts of GS [Hertel et al., 2022]. The characterization of GS-resistant Burkholderia anthina mutants uncovered a novel GS resistance mechanism, which is based on the overproduction of PEP [Schwedt et al., 2023a]. Similar to B. anthina, the EPSP synthase of B. subtilis does not permit amino acid exchanges due to a high demand for shikimate pathway intermediates [Schwedt et al., 2023b]. The evolution of GS resistance allows to elucidate the peculiarities of the shikimate pathway in B. subtilis.
Selected publications
Riedel R, Commichau FM, Benndorf D, Hertel R, Holzer K, Hoelzle LE, Mardoukhi MSY, Noack LE, Martienssen M (2024) Biodegradation of selected aminophosphonates by the novel bacterial isolate Ochrobactrum sp. BTU1. Microbiol Res. 280:127600.
Schwedt I, Schöne K, Eckert M, Pizzinato M, Winkler L, Knotkova B, Richts B, Hau JL, Steuber J, Mireles R, Noda-Garcia L, Fritz G, Mittelstädt C, Hertel R, Commichau FM (2023b) The low mutational flexibility of the 5-enolpyruvyl-shikimate-3-phosphate synthase in Bacillus subtilis is due to a higher demand for shikimate pathway intermediates. Environ Microbiol. 25: 3604 - 3622.
Schwedt I, Collignon M, Mittelstädt C, Giudici F, Rapp J, Meißner J, Link H, Hertel R, Commichau FM (2023a) Genomic adaptation of Burkholderia anthina to glyphosate uncovers a novel herbicide resistance mechanism. Environ Microbiol Rep. 15: 727-739.
Hertel R, Schöne K, Mittelstädt C, Meißner J, Zschoche N, Collignon M, Kohler C, Friedrich I, Schneider D, Hoppert M, Kuhn R, Schwedt I, Scholz P, Poehlein A, Martienssen M, Ischebeck T, Daniel R, Commichau FM (2022) Characterization of glyphosate-resistant Burkholderia anthina and Burkholderia cenocepacia isolates from a commercial Roundup solution. Environ Microbiol Rep 14: 70-84.
Hertel R, Gibhardt J, Martienssen M, Kuhn R, Commichau FM (2021) Molecular mechanisms underlying glyphosate resistance in bacteria. Environ Microbiol 23: 2891-2905.
Wicke D, Schulz LM, Lentes S, Scholz P, Poehlein A, Gibhardt J, Daniel R, Ischebeck T, Commichau FM (2019) Identification of the first glyphosate transporter by genomic adaptation. Environ Microbiol 21: 1287-1305.
