Molecular evolution
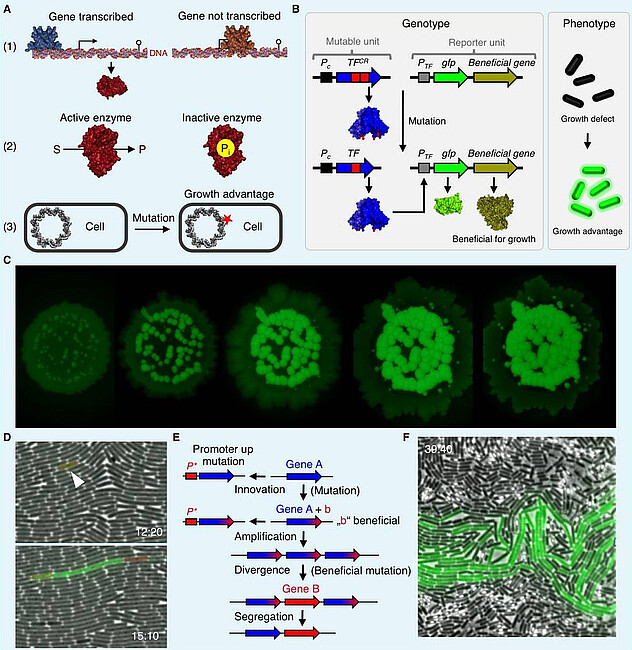
The accumulation of beneficial mutations (genomic adaptation) is essential for survival of an organism if it is unable to adapt to the environment by adjusting gene expression or metabolism accordingly. Mutations such as nucleotide exchanges, deletions and insertions are crucial for the emergence and evolution of proteins with novel functions, which is also a prerequisite for the origin of novel species, thus for the diversity of life. However, the mutation rate has to be kept low to prevent the accumulation of harmful mutations that appear more often. Tandem repeats (TRs) are mutational hot spots that are present in the genomes of all living organisms. The modification of TRs due to the deletion or the addition of repeat units can account for severe human disorders. Moreover, TRs can cause genome instability of bacterial strains that were engineered for biotechnological purposes. To understand the emergence of TR-related human diseases it is important to elucidate the molecular mechanism underlying TR mutagenesis. Moreover, the identification and elimination of factors that are involved in TR mutagenesis may help to create stable production strains for industrial applications. Microorganisms such as the Gram-positive and Gram-negative bacteria Bacillus subtilis and Escherichia coli, respectively, are well-suited model systems to study genomic adaptation because they grow fast, they are easy to handle and propagate, their cultures reach high cell densities, they are exceedingly well studied, and whole-genome sequencing techniques are cheap and well established. However, despite these advantages of using bacteria to address evolutionary questions and to elucidate the molecular mechanisms underlying genomic adaptation there are also drawbacks. For instance, to determine the speed and time course of a mutation to occur, phenotypes of many individuals from large cell populations have to be compared and statistical analyses are required. Moreover, single or multiple genomic alterations that occur at different loci often cause the same phenotype. Therefore, it is difficult to infer the genotype of an evolved organism from its phenotype. We are interested in the evolution of bacteria by genomic adaptation due to gene amplification, single nucleotide exchanges, insertions and deletions. To study bacterial evolution, we develop mutation reporter systems that is suitable to visualize mutational events such as deletions and gene (de)amplification in B. subtilis at the level of single cells. Applying the mutation reporter system in combination with microfluidics will be helpful to elucidate the molecular mechanism underlying genomic (in)stability in bacteria. Moreover, the approach can be applied to assess whether the selective pressure that is acting on the cell population affects the mutation rate.
Selected publications
Richts B Lentes S Poehlein A Daniel R Commichau FM (2021) A Bacillus subtilis pdxT mutant suppresses vitamin B6 limitation by acquiring mutations enhancing pdxS gene dosage and ammonium assimilation. Environ Microbiol Rep. 13: 218-233.
Dormeyer M Lentes S Ballin P Wilkens M Klumpp S Kohlheyer D Stannek L Grünberger A Commichau FM (2018) Visualization of tandem repeat mutagenesis in Bacillus subtilis. DNA Repair (Amst) 63: 10-15.
Dormeyer M Lübke AL Müller P Lentes S Reuß DR Thürmer A Stülke J Daniel R Brantl S Commichau FM (2017) Hierarchical mutational events compensate for glutamate auxotrophy of a Bacillus subtilis gltC mutant. Environ Microbiol Rep 9: 279-289.
Dormeyer M Egelkamp R Thiele MJ Hammer E Gunka K Stannek L Völker U Commichau FM (2015) A novel engineering tool in the Bacillus subtilis toolbox: inducer-free activation of gene expression by selection-driven promoter decryptification. Microbiology 161: 354-61.
Gunka K Stannek L Care R Commichau FM (2013) Selection-driven accumulation of suppressor mutants in Bacillus subtilis: the apparent high mutation frequency of the cryptic gudB gene and the rapid clonal expansion of gudB+ suppressors are due to growth under selection. PLoS One 8: e66120.
Gunka K Tholen S Gerwig J Herzberg C Stüke J Commichau FM (2012) A high-frequency mutation in Bacillus subtilis: requirements for the decryptification of the gudB glutamate dehydrogenase gene. J Bacteriol 196: 1036-1044.
