Amino acid metabolism in bacteria
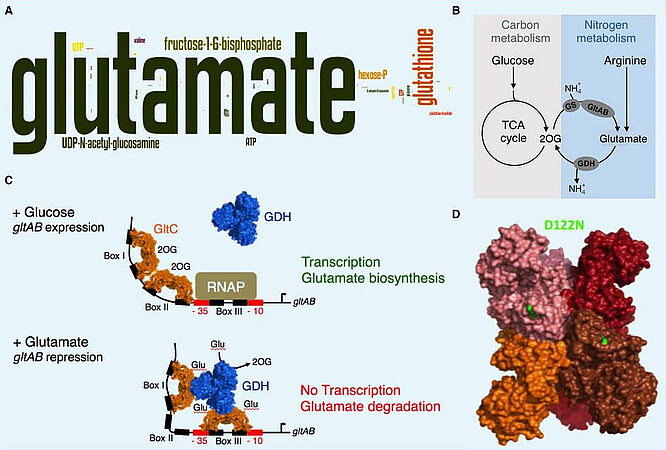
Glutamate and glutamine are the major amino group donors for all nitrogen-containing compounds in a cell (Figure A). We are interested in glutamate metabolism in the Gram-positive model bacterium Bacillus subtilis. The enzymatic reactions involved in the synthesis and degradation of glutamate represent a central metabolic node, linking carbon to nitrogen metabolism (Figure B). B. subtilis relies on the glutamine synthetase (GS) and the glutamate synthase (GltAB) for biosynthesis of glutamate from ammonium and 2-oxoglutarate (2OG). The glutamate dehydrogenases (GDHs) of B. subtilis are strictly catabolically active. During growth of B. subtilis with glucose and ammonium the transcription of the GltAB encoding gltAB genes is activated by GltC (Figure C). Under these growth conditions, the GDH encoding rocG gene is repressed. During growth with nitrogen sources like arginine, which is converted to glutamate and induces the expression of the rocG gene, the glutamate levels increase and the gltAB genes are not transcribed (Figure C). The GDH RocG degrades glutamate to ammonium and 2OG, and prevents the transcription factor GltC from activating transcription of the gltAB genes. The enzyme forms a transient complex with the transcription factor GltC via a direct protein-protein interaction. The GDH RocG is a trigger enzymes are active in metabolism and in controlling gene expression. Trigger enzymes form a subgroup of moonlighting proteins (http://www.moonlightingproteins.org). A single amino acid exchange is sufficient to convert the bifunctional GDH RocG into a monofunctional enzyme that is solely active in metabolism (Figure D). We have also demonstrated that the active variant GudB1 of the paralogous GDH GudB, which is present in undomesticated B. subtilis isolates, can replace RocG. Thus, B. subtilis possesses two GDHs that are active in glutamate degradation and in controlling de novo synthesis of glutamate. This sophisticated regulatory system enables the bacteria to adjust glutamate biosynthesis and degradation depending on the available carbon and nitrogen sources. The importance of glutamate homeostasis is underlined by the fact that B. subtilis responds to mutations interfering with glutamate metabolism by rapidly accumulating extragenic or intragenic suppressor mutations to bring the glutamate supply into balance. Recent genetic data indicate that either RocG or GudB convert the transcriptional GltC into a repressor of the gltAB genes (Figure C). Currently, we are elucidating the molecular details of the ternary RocG-GltC-DNA and GudB1-GltC-DNA complexes. We also work on the identification of novel amino acid transporters in B. subtilis.
Selected publications
Völker F, Maaß S, Phan ANT, Gibhardt J, Commichau FM Blank LM (2025) High glutamate demand enables simultaneous consumption of glycerol and citrate despite carbon catabolite repression in engineered Bacillus subtilis strains. Metab Eng. 91: 379-388.
Mardoukhi MSY, Rapp J, Irisarri I, Gunka K, Link H, Marienhagen J, de Vries J, Stülke J, Commichau FM (2024) Metabolic rewiring enables ammonium assimilation via a non-canonical fumarate-based pathway. Microb Biotechnol. 17: e14429.
Meißner J, Königshof M, Wrede K, Warneke R, Mardoukhi MSY, Commichau FM, Stülke J (2024) Control of asparagine homeostasis in Bacillus subtilis: identification of promiscuous amino acid importers and exporters. J Bacteriol. 206: e00420-23.
Dormeyer M, Lentes S, Richts B, Heermann R, Ischebeck T, Commichau FM (2019) Variants of the Bacillus subtilis LysR-type regulator GltC with altered activator and repressor function. Front Microbiol. 10: 2321.
Reuß DR, Rath H, Thürmer A, Benda M, Daniel R, Völker U, Mäder U, Commichau FM, Stülke J (2018) Changes in DNA topology affect the global transcription landscape and allow rapid growth of a Bacillus subtilis strain lacking carbon catabolite repression. Metab Eng 45: 171-179.
Dormeyer M, Lübke AL, Müller P, Lentes S, Reuß DR, Thürmer A, Stülke J, Daniel R, Brantl S, Commichau FM (2017) Hierarchical mutational events compensate for glutamate auxotrophy of a Bacillus subtilis gltC mutant. Environ Microbiol Rep. 9: 279-289.
Stannek L, Thiele MJ, Ischebeck T, Gunka K, Hammer E, Völker U, Commichau, FM (2015) Evidence for synergistic control of glutamate biosynthesis by glutamate dehydrogenases and glutamate in Bacillus subtilis. Environ Microbiol. 17: 3379-3390.
Stannek L, Gunka K, Care RA, Gerth U, Commichau FM (2015) Factors that mediate and prevent degradation of the inactive and unstable GudB protein in Bacillus subtilis. Front Microbiol. 7: 758.
Stannek L, Egelkamp R, Gunka K, Commichau FM (2014) Monitoring intraspecies competition in a bacterial cell population by co-cultivation of fluorescently labelled strains. J Vis Exp 18: e51196.
Gunka J, Stannek L, Care RA, Commichau FM (2013) Selection-driven accumulation of suppressor mutants in Bacillus subtilis: the apparent high mutation frequency and the rapid clonal expansion of gudB(+)suppressors are due to growth under selection. PLoS One 8: e66120.
Gunka K, Commichau FM (2012) Control of glutamate metabolism in Bacillus subtilis: A complex interplay between ammonium assimilation, glutamate biosynthesis and degradation. Mol Microbiol. 85: 213-224.
Gunka K, Gerwig J, Tholen S, Herzberg C, Stülke J, Commichau FM (2012) A high frequency mutation in Bacillus subtilis: decryptification of the gudB glutamate dehydrogenase gene. J Bacteriol 194: 1036-1044.
Gunka K, Newman J, Commichau FM, Herzberg C, Rodrigues C, Hewitt L, Lewis R, Stülke J (2010) Functional dissection of a trigger enzyme: Mutations of the Bacillus subtilis glutamate dehydrogenase RocG that affect differentially its catalytic activity and regulatory properties. J Mol Biol. 400: 815-827.
Commichau FM, Stülke J (2008) Trigger enzymes: bifunctional proteins active in metabolism and in controlling gene expression. Mol Microbiol. 67: 692-702.
Herzberg C, Flórez Weidinger LA, Dörrbecker B, Hübner S, Stülke J, Commichau FM (2007) SPINE: A method for the rapid detection and analysis of protein-protein interactions in vivo. Proteomics 12: 4032-4035.
Commichau FM, Herzberg C, Valerius O, Tripal P, Stülke J (2007) A regulatory protein-protein interaction governs glutamate biosynthesis in Bacillus subtilis: The glutamate dehydrogenase RocG moonlights in controlling the transcription factor GltC. Mol Microbiol. 65: 642-654.
Commichau FM, Wacker I, Schleider J, Blencke HM, Reif I, Tripel I, Stülke J (2007) Characterization of Bacillus subtilis mutants with carbon source-independent glutamate biosynthesis. J Mol Microbiol Biotechnol 12: 106-113.
Commichau FM, Forchhammer K, Stülke J (2006) Regulatory links between carbon and nitrogen metabolism. Curr Opin Microbiol. 9: 167-172.
